The written version of our genetic instruction manual, which has 3 billion letters, would take up many volumes’ worth of space in real life. However, it is only contained inside the tiny cellular structure of our body. From our gender to our physical characteristics to our susceptibility to disease, practically every aspect of our lives is determined by the choices our ancestors made. However, for a very long time, no one knew what this genetic code looked like and what it contained. Scientists eventually uncovered the shape, language, and exact function of our DNA, with some unexpected findings along the way.
The genetic specifications for all known creatures and many viruses are stored in a deoxyribonucleic acid or DNA, a polymer made up of two polynucleotide chains that coil around each other to create a double helix. DNA governs development, functioning, growth and reproduction.
Two men did change the world of science

In a statement made 70 years ago, according to James Watson, only a few discoveries have been of such exquisite beauty as DNA. Watson was referring to the double helix, a structure that is 2.5 nanometers in diameter, looks like a helically twisted rope ladder, and stands 7,2 feet (2,2 meters) in length if fully unfolded.
On April 25, 1953, James Watson and Francis Crick published a single page in Nature proposing a model for the three-dimensional structure of deoxyribonucleic acid (DNA), the molecule that encodes human genes.
It seemed that the two researchers were confident in the long-term relevance of their model since they cited “novel features of considerable biological interest” at the start of their paper.
Zero interest in chemistry
Although at first glance, it did not seem likely that two “scientific clowns,” as scientist Erwin Chargaff dubbed them, would produce such a groundbreaking discovery. James Watson, who was very talented, began studying biology at the University of Chicago when he was only 15 years old. Birds were his major focus at the time, thus he was able to avoid taking any science classes.
The zoologist’s understanding of chemistry and physics was quite limited when he first arrived at the Cavendish Laboratory in Cambridge, England, in the autumn of 1951, at the tender age of 23. In England, he met British scientist Francis Crick, who was 13 years older and whose loud laughing was the bane of his colleagues’ existence. Francis Crick’s prior life as a researcher was summed up by the institute’s director, Sir Lawrence Bragg. According to him, Francis was talking ceaselessly and had come up with next to nothing of decisive importance.
A scientific footrace
In 1949, Erwin Chargaff discovered that the DNA bases adenine, thymine, cytosine, and guanine always occur in DNA at a 1:1 ratio, or most likely in pairs. The next step was to figure out the structural integrity of the bases and how they fit together. Watson, who was originally uninterested, attended a presentation by neighboring King’s College London scientist Rosalind Franklin in November 1951, during which she shared recent X-ray diffraction photographs of DNA.
Waston found intriguing her speculation that DNA could exist in a twisty helical shape with two, three, or four twists. As soon as Watson and Crick got back to Cambridge, they set out to try to replicate this structure. They hypothesized, based on chemical calculations, that the structure would consist of three chains joined in a helix by magnesium ions, with the molecular arms pointing in all directions.
Success through failure
Watson, however, was not paying close attention, and the team’s model of the chemical reaction turned out to be incorrect. They made a disappointing appearance in front of Rosalind Franklin and London-based biophysicist Maurice Wilkins.
Colleagues were quite harsh in their criticism. Previous X-ray images produced by these two scientists had demonstrated conclusively that the supporting chains could not lay within, refuting the premise of Watson and Crick, and that magnesium ions were scarcely capable of maintaining this structure.
In July of 1952, Erwin Chargaff visited Watson and Crick in the lab and delivered a similarly damning assessment of their scientific prowess: “enormous ambition and aggressiveness, coupled with an almost complete ignorance of, and a contempt for, chemistry…”
When it became public that famous scientist Linus Pauling, on the other side of the Atlantic, shared a fascination with the structure of genetic information and suggested a model for it, the scientific reaction intensified. Urgency necessitated swift action.
Then, towards the end of 1952, Maurice Wilkins offered Watson and Crick an X-ray structural study of his colleague, Rosalind Franklin, which proved to be a pivotal event that ultimately led to triumph. It was, in fact, a picture of a recently discovered DNA structure. But this was all without Franklin’s consent.
By the end of the study, the two scientists had reached a consensus: DNA is made up of two strands that intertwine with each other like rungs on a rope ladder. Hydrogen bonds hold their molecular appendages, the complementary bases, together. Finally, Watson and Crick assembled their metal double helix structure like pieces of a jigsaw. This variation won over even the most skeptical people.
Recognition and respect
Many scientists date the beginning of molecular genetics to the publication of the “Watson-Crick Model” of the structure of DNA. James Watson, Francis Crick, and Maurice Wilkins all split the 1962 Nobel Prize in Medicine and Physiology equally. In contrast, Rosalind Franklin, whose research offered the last vital piece of the puzzle, came up empty. Sadly, she passed away from uterine cancer in 1958, when she was only 37 years old, without seeing the fruits of her labor. It’s safe to say that most people nowadays have forgotten who Franklin and Wilkins were. But the names Watson and Crick will forever be linked to the double helix model of DNA.
The exchange of information
Translation of genetic code
Since 1953, scientists have understood the fundamental nature of our DNA and that it includes the blueprints for every aspect of our identities, from physical appearance to health. It was quickly understood that each base pair represented a different letter in the manual. The question is how to give form to these inscrutable directives and create a live, breathing, and authentic human being.
In every human cell, two sets of 23 chromosomes are created when an egg from the mother and a sperm from the father fuse. The maternal contribution to these roughly X-shaped structures is half, whereas the paternal contribution is half.
We store and carry our DNA, or genetic information, in a compact form called chromosomes. All of our DNA, together with its protective envelope structures, is stored in a very condensed form on the many chromosomes in our bodies.
The sequence of bases as the alphabet of life
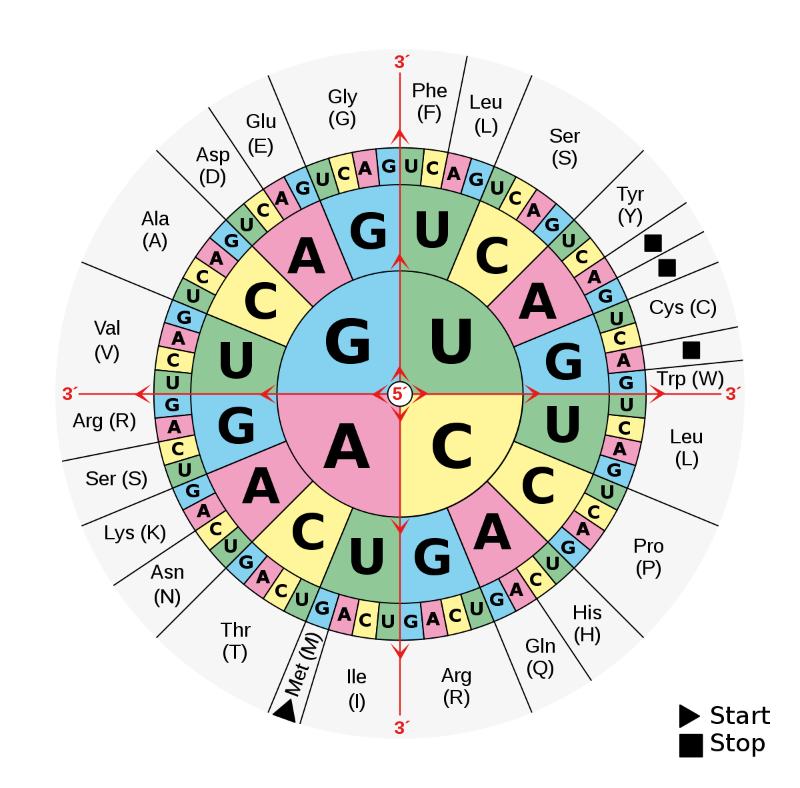
Adenine (A), Guanine (G), Thymine (T), and Cytosine (C) are the four bases that make up DNA. The two strands of DNA’s framework are held together by the pairs A-T and C-G. They serve as the rungs on this hereditary ladder. When you align the ladder segments of a single DNA strand, you’ll see a lengthy string of base letters.
And it is in them where the genetic code is found. Three of these letters are put together to spell out a word that specifies where in the process the production of protein should include a certain amino acid. A string of these letters forms a phrase, which in turn becomes the blueprint for a protein. And these molecules, in turn, play the role of a biochemical housekeeper who makes sure everything from cell and tissue formation to signal transduction and metabolic processes go smoothly.
Transcription and translation of DNA
Genes and proteins
How, however, does the blueprint for a structure end up as a protein? The process of making proteins from scratch is called biosynthesis. At this phase, the genome must be unpacked so that the information for a protein can be read from DNA. Generally found as a double strand, DNA separates into two single strands. Thus, the free arms of the rope ladder become accessible.
Beginning with the copy
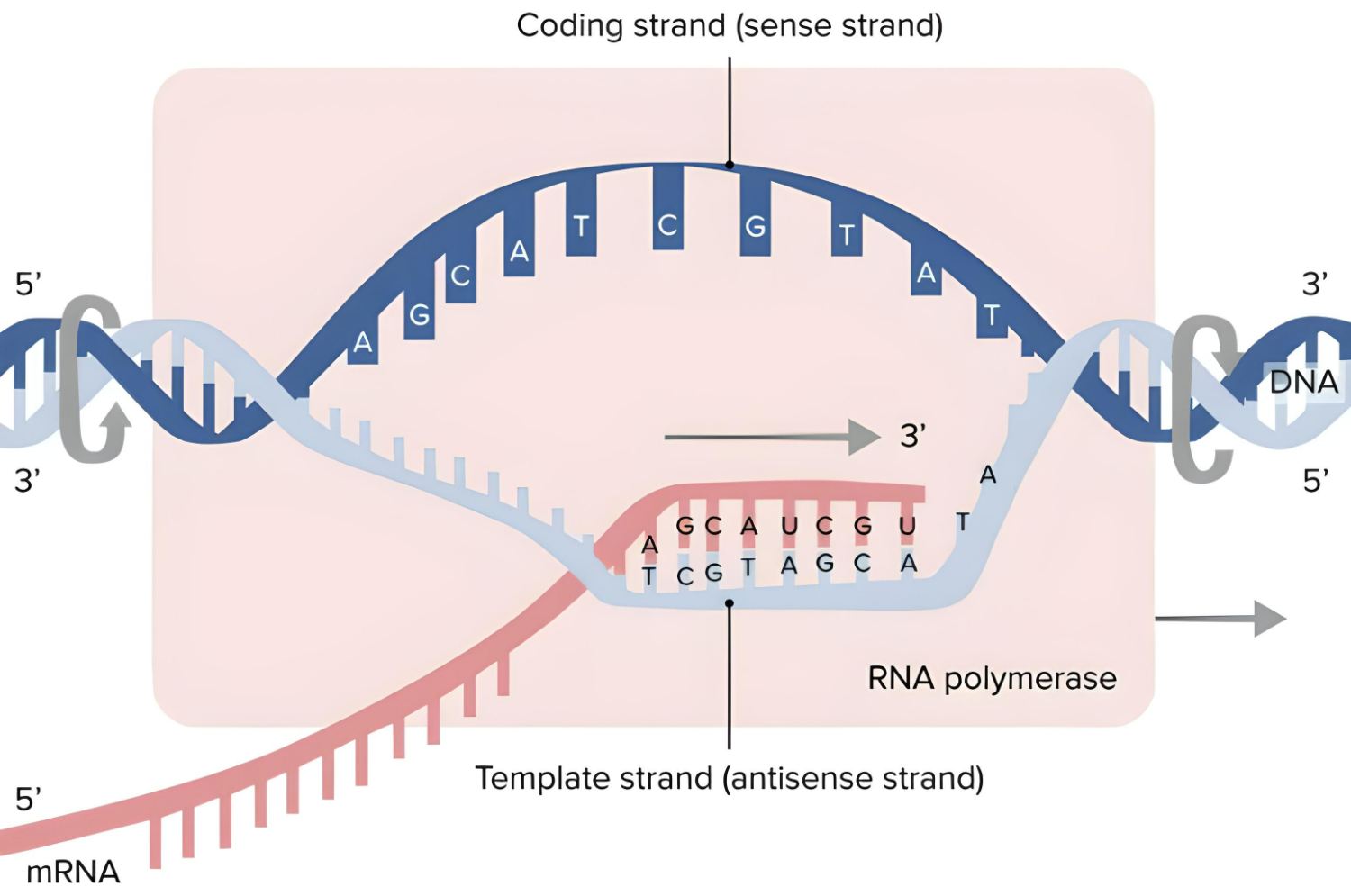
Enzymes have now made it possible to make carbon copies of this segment of the strand by simply joining a complementary base to each of the free arms. This time around, though, the base uracil bonds to the adenine instead of the thymine. In this case, however, the ribonucleic acid (RNA) serves as the scaffolding for these newly joined bases. After the copy is complete, enzymes cut the RNA strand and its accompanying DNA bases away, creating a copy of this region of the genome that can be moved throughout the cell, the messenger RNA (mRNA). Transcription refers to this process of making a copy of the genetic material and rewriting it.
Translation into protein building blocks
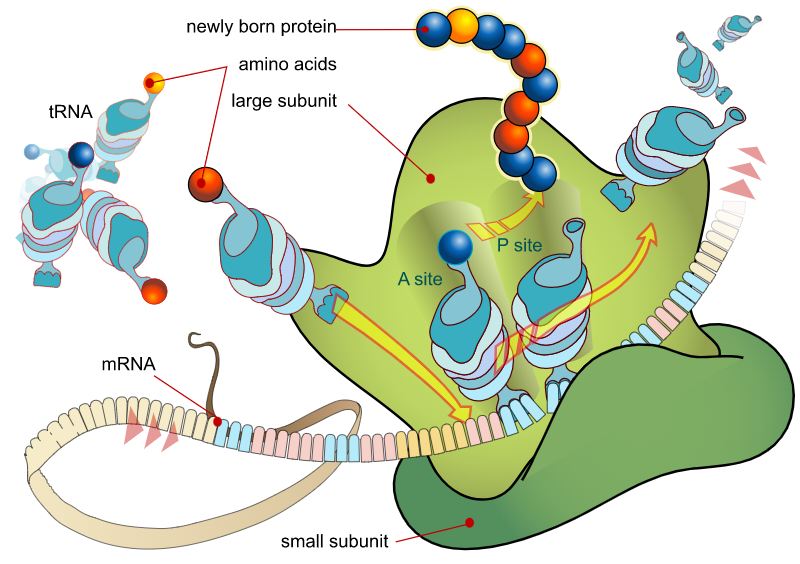
However, this is just the beginning. mRNA now transports the genetic information copy from the nucleus into the cell plasma, where it will be read by the ribosomes and used to make proteins.
The mRNA is sandwiched between the two subunits, one larger than the other; this creates a reading unit similar to the needle on a tape recorder. It decodes the genetic code by identifying which of the three bases (and hence which genetic code word) is present in each instance.
Simultaneously, many of the amino acids that will make up the future protein accumulate on the ribosomes, each of which has a tiny piece of RNA consisting of precisely three base letters connected to it. These letters and numbers serve as a label, identifying the specific amino acid bound to this transport RNA (tRNA). It is the job of the ribosome to dock and connect the component of the amino acid that corresponds to the coding of the next amino acid in the read-out mRNA.
Polypeptides, or chains of amino acids, are produced in this fashion and are the building blocks from which proteins are assembled. DNA can only perform its job via translation, the process by which the genetic information is converted into a chain of amino acids.
Junk DNA
Discarded material transformed into a control center
A gene is a set of instructions for making a particular protein; it consists of a specific sequence of the base pairs cytosine, adenine, guanine, and thymine. It’s the blueprint for these critical messengers of our body’s processes.
It was quickly discovered, however, that significant portions of human DNA lacked any recognizable construction instructions. Sequences in an organism’s DNA that do not code for proteins are known as noncoding DNA (ncDNA). They looked to be made up of illogical and repeated DNA sequences that had no discernible purpose. Therefore, scientists called these pieces of DNA “junk DNA.”
Only 2% of your genes are real
However, scientists were baffled when they looked more closely at the breakdown of our genetic material and saw that around 44% of it is “junk” in the form of several copies of genes and gene fragments (repeats).
In addition, 52% seems to be useless as well and does not code for proteins. However, only around 2%–4% of human DNA is made up of genuine protein-coding genes.
It has long been a puzzle as to why evolution has preserved so much irrelevant DNA in addition to these gene sequences. But this issue was first answered by research in 2004. Scientists in the United States revealed a startling discovery about this “living genome deserts” that many regions of DNA that do not code for proteins were far from inactive. They include sequences that may activate or silence other genes, even if they are located far away.
A regulator made of “junk”?
This suggests that the genome’s so-called “junk” is playing a significant role in regulating gene activity, helping to shed light on the basic differences across species even though their genes are, on average, just a few percent different.
Also, scientists from LLNL and JGI found that different parts of junk DNA have experienced different degrees of modification during evolution. There are several non-coding regulatory elements in the “desert areas” that are resistant to rearrangement and defend themselves via repeating junk DNA patterns. It appears that genomic regions known as stable genome deserts are essentially hidden gene regulatory components that preserve the intricate function of neighboring genes.
About two-thirds of the genome deserts and about 20% of the overall genome could be gene segments that are completely useless for biology, indicating that most of the genome is redundant. At least, 75 percent or more of our genetic material is really just junk and only around 8–14% of our DNA is functional in some way.
Our genome is governed by junk DNA
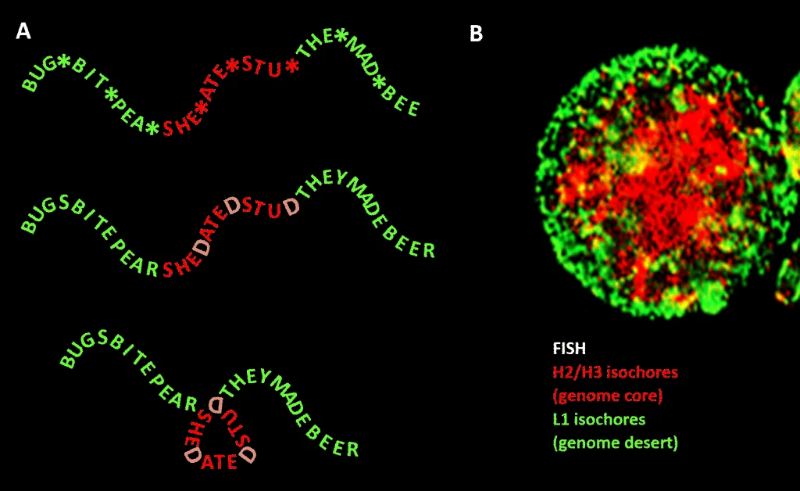
The term “junk DNA” refers to the 98% of the human genome that does not code for proteins but the truth is actually more complicated.
Because this notion of mostly useless junk DNA kind of shattered in 2011. The international ENCODE project discovered something astounding: almost all of our junk DNA functions as a massive control panel for our genome, containing millions of molecular switches that can activate and deactivate our genes as needed, including in regions where only an “unstable desert” had been suspected.
The “junk” has millions of switches
Scientists created a detailed map of the locations and distributions of control elements, which revealed that the control switches are often located in inconveniently distant genomic regions from the genes they regulate. However, due to the complex three-dimensional shape of the DNA strands, they can still come close and exert their regulatory effects.
That means, our genome is only functional because of switches: millions of buttons that control which genes are active.
Genes derived from junk DNA
But junk DNA has other, non-regulatory functions too; scientists from Europe identified a gene on mouse chromosome 10 that appeared out of nowhere but originated between 2.5 and 3.5 million years ago via genome-wide comparisons. The gene was the only one positioned in the center of a lengthy non-coding chromosomal part. This area is present in all other mammalian genomes as well. However, the gene is only found in mice.
There was some speculation that a gene may emerge at a place in the genome that had never been used before, but no evidence for this had ever been found. Yet it was discovered that the mutations that only occur in mice could be responsible for the new formation of the gene.
This demonstrates that the regions of DNA that do not code for proteins are an essential component of our genome and that they have long played a significant role in a variety of modern genetic analyses.
DNA and forensic science
It was all solved via a DNA analysis

You can identify a criminal by his genetic fingerprint from as little as a drop of saliva on a Coke bottle or cigarette filter, a few skin cells beneath the victim’s fingernails, or blood on his clothes.
Most of our bodily fluids also contain cells from our body, and with them our genetic information. Skin cells are always left behind when a hand is dragged along a rough surface or is scratched, and these cells carry our DNA.
However, there is a catch: there is far too little genetic material in the crime scene that remains for analysis, and this is precisely the reason why DNA analyses from such relics were unattainable for a long time. These genetic material remains can tell investigators whether or not their suspect was the perpetrator.
The polymerase chain reaction for DNA testing
But in 1983, US scientist Kary B. Mullis came up with a plan to multiply the few amounts of DNA that obtained and, in the process, devised one of the most pivotal techniques in genetics and biotechnology: the polymerase chain reaction (PCR).
A DNA fragment of up to 3,000 base pairs in length is heated to 201 to 205 degrees Fahrenheit (94 to 96 degrees Celsius), which breaks the hydrogen bonds between the bases of the double strand, resulting in the separation of the helix into two single strands. Two primers are then added to the DNA solution.
They bind to certain places on the DNA segments (based on their structures) and signal the beginning of the copying process, which is carried out by a heat-stable enzyme called polymerase.
At a temperature of 140–160 degrees Fahrenheit (60–70 degrees Celsius), it joins DNA-building components floating in solution to produce a perfect replica of the sequence designated by the primers, leading to another double strand and doubling the original amount of sequences.
Once the PCR is finished, the few remnants from the murder scene become a solution containing millions of copies of the perpetrator’s DNA, thanks to the process of repeated cycles in which the double strands are split from one another and then supplied with new halves by the polymerase.
A unique repeat pattern
The testing phase can now begin, with researchers comparing only small fragments of the DNA rather than the full sequence (which would take too long and be too laborious).
These fragments are found in the genome’s non-coding regions and are made up of several repeating base sequences termed “short tandem repeats” (STRs), which provide a unique genetic fingerprint since their numbers vary from person to person.
In many countries, a standard DNA analysis at a criminal lab includes testing for eight STR systems over several chromosomes and one sex-differentiating characteristic, which should be more than enough to rule out the possibility of a chance match.
Estimates suggest that the number of people with whom our unique STR pattern is shared is less than one in a billion, with the exception of identical twins. If a suspect’s genetic fingerprint matches that found at the crime scene, then it’s likely that s/he committed the crime in question; her/his own DNA has in fact convicted him.
Probing the paternity of a child
The mother’s identity is generally evident since she gives birth to the kid (barring surrogate moms), but the identity of the father is not always so clear.
It’s possible that the question of paternity won’t come up until the child is an adult if the woman has cheated on her partner in secret or if she gets pregnant shortly before breaking up with her partner and keeps the baby from him.
Numerous laboratories around the world have long offered such gene-based paternity tests online, and the process for those willing to take the test is very simple: just send in a saliva sample, a few hairs with a hair root, a baby’s pacifier covered with spit, or a piece of chewing gum that has been well chewed.
First, the DNA is extracted from the samples and amplified by polymerase chain reaction (PCR) in the lab; next, the DNA is compared to samples of the same genetic material from the child’s father or, ideally, the mother.
Short tandem repeats (STRs) are also used in forensic DNA analysis, and the frequency with which a given base sequence is repeated within an STR marker varies from person to person but is passed down from parents to offspring. Each person carries two STR marker variants at each gene locus, one inherited from mother and one from father.
In contrast, if the genetic material of the parent and child differs at three or more STR markers, paternity or maternity is considered to be ruled out. The probability that two unrelated people will have the exact same pattern of repeats at these markers is just one in 100 billion, according to current estimates.
The Human Genome Project (1990-2003)
Learning by reading life’s book
In the year 2000 in the United States, Bill Clinton and his British counterpart, Tony Blair, arranged for an unusual news conference in Washington. Nothing less than the human DNA itself was at stake here. The decoding of our genetic composition has been publicly announced by Clinton and, following him, by representatives of two rival research organizations, one government and one private.
And in 2022, scientists finally announced that they finished decoding the entire human genome. According to that, about 30,000 human genes are housed in the nucleus of each human cell, where they are contained in 23 chromosomal groups.
Humanity’s next big thing
An early version of the “Book of Life” has been deciphered by both the worldwide Human Genome Project (HGP) scientists and genetic engineering pioneer Craig Venter and his business Celera. About 3.1 billion letters make up our genome, which is composed entirely of apparently random sequences of the four nucleotide bases (adenine, cytosine, guanine, and thymine).
From the neurons that carry impulses throughout the brain to the immune cells that help protect us from external attack, each of the trillions of cells that make up our bodies has the same 3.1 billion DNA base pairs that make up the human genome.
It’s still not fully known what words and sentences may be constructed from these letters, as well as where certain functional units of genetic material are buried.
The decipherment of the human genome paved the way for novel approaches to illness prevention, diagnosis, and treatment. But these 3.1 billion letters of sequence in one human DNA were only the beginning of the long road to deciphering the human genome.
Interesting, but impossible
Things looked very different 25 years ago. In 1985, a group of genetics experts at the University of California, Santa Cruz, were approached by biologist Robert Sinsheimer with an unusual proposal: Why not try to sequence the human genome? The response was as unanimous as it was unequivocal: bold, exciting, but simply not feasible. Decoding even small sections of DNA was still too laborious at this time.
However, one of the researchers involved, Walter Gilbert of Harvard University, did not give up on the idea. About 20 years ago, he and a colleague were the first to develop a method for reading out the genetic code or genetic sequencing.
However, potential backers were still cautious, asking, “What if it turns out that the entire thing is not worth the massive effort?” and “Shouldn’t we possibly start with the genome of a small, less sophisticated creature, such as a bacterium?”
Genome arms race
Finally, in 1988, the U.S. National Institutes of Health (NIH) was convinced to organize a project to decode the human genome, led by none other than James Watson, one of the two discoverers of the double helix structure of DNA.
Understanding of the disease genes
However, progress has been sluggish since researchers were always debating whether or not it would be more efficient to begin by searching for illness genes rather than meticulously sequencing everything.
Craig Venter, a geneticist at the National Institute of Neurological Disorders and Stroke (NINDS), stood out because he and his colleagues had created a novel approach to discover gene fragments at an unparalleled rate, but without understanding their function.
Watson opposed and publicly complained about the sellout of genetic material which was met with early enthusiasm by NIH leaders since, if patented, these genes could be converted into cash. The fallout was seen when Watson was replaced as project head by Francis Collins in April 1992.
Not fast enough
Collins made a dismal prediction in 1993 that human genome sequencing wouldn’t be finished until 2005 at the earliest if things kept moving at their current rate. Part of the reason for this was the lack of resources that have so far prevented the development and widespread use of state-of-the-art DNA sequencers, which would greatly facilitate the automation of the genome decoding process.
On the other side, achieving a success rate of 99.99 percent was a need. After all, international research institutes were joining the effort at an increasing rate.
Upon meeting Craig Venter in 1995, the HGP researchers and management were rudely roused. As part of his new job at a commercial corporation, Venter released the first genome of a fully developed organism, that of the Haemophilus influenzae bacteria. He had accomplished it in a year because of the cutting-edge computing power available at the time. While progress was being made by Collins and the HGP researchers, it was slower than some would like.
Head to head
At the 1998 annual gathering of genetic experts, Venter pulled off his next move by announcing that his new firm would be able to decode the human genome in three years for a quarter of the cost of the HGP. He would be assisted by an automated sequencing system currently under development.
At this point, Collins and his group must take action. Six months later, they announced that instead of waiting until 2005, full genome sequencing was now expected to be completed in 2003 thanks to increased efforts. They wanted to provide the first functional version of the human genome that is around 90% valid by spring 2001.
It seemed like Craig Venter and his business, Celera, were in for a close finish. In reality, though, efforts to establish a mutually agreeable human genome resolution had already begun behind the scenes.
The HGP suggested holding a combined press conference to announce the initial versions of both projects at the same time on July 26, 2000. While the HGP had been publishing their sequencing in the British journal “Nature,” Venter and his colleagues had been contributing to the rival American journal “Science.” The unveiling of the virtually entire human genome was announced two years ahead of schedule, on April 14, 2003.
Thus, in April 2003, the Human Genome Project (HGP) was announced as completed but only around 85% of the genome was actually included. 15% of the remaining human genome was sequenced only by January 2022.
Dictionary of genetics
Amino acids
20 amino acids are the fundamental building blocks of proteins, and the genes determine the order in which these amino acids are put together to create a chain.
Bases
The nucleotides adenine (A) and thymine (T) and cytosine (C) and guanine (G) are paired with one another in double-stranded DNA through the complementary base pairing concept. Thymine (T) is switched out for uracil (U) in the ssRNA (single-stranded RNA).
Chromosomes
Chromosomes, which contain an organism’s genetic information, number 46 in humans thanks to the duplication of the 23 chromosomes found in each of our cells.
Codon
An amino acid’s genetic code is encoded in a sequence of three bases.
DNA
Deoxyribonucleic acid is a double-stranded molecule composed of a sugar backbone (deoxyribose) and a phosphate group, and a linear series of base pairs. The two single strands are complementary to each other, run in an antiparallel direction, and are kept together by base pairs.
DNA sequence
An order of the DNA molecule’s construction order.
Dolly
A well-known cloned sheep that was cloned in 1996 from an adult sheep’s single cell.
Gene
Genes are sections of DNA. In eukaryotes, genes are often made up of coding sections (exons) and noncoding sections (introns). Coding portions (exons) carry the genetic information for creating proteins or functional RNA (e.g., tRNA).
Genetics
Molecular genetics investigates the fundamental laws of heredity at the molecular level, whereas classical genetics focuses on the inheritance of characteristics, especially in higher species. Applied genetics focuses on the breeding of economically highly productive crops and animals.
Genetic code
The genetic code is a kind of encryption used to store information on DNA, and it is represented by a set of three-base pairs in all known forms of life.
Genetic fingerprint
Genetic fingerprints are unique to each person and are generated by using so-called restriction enzymes and undergoing further analytical processes.
Genome
A genome refers to the whole set of genetic instructions for a certain organism.
Human Genome Project
An international effort funded by many agencies to investigate the DNA sequence, protein function, and regulatory mechanisms of the human genome.
Gamete
Gametes are sexually reproducing cells (eggs, sperm) that contain just one copy of each of the 23 genes found in the human genome, which is called haploid (in humans).
Cloning
Producing offspring with the same genetic material by cell division or nuclear transplantation.
Mutation
Mutations, which may be caused by anything from exposure to ultraviolet light or naturally occurring radioactivity to the simple passage of time, are the fundamental mechanism by which new species are created and evolve.
Nucleic acids
Both DNA and/or RNA
Nucleotides
A phosphate group, a sugar, and a base make up the three components of the DNA-building block.
Nucleus
The nucleus is the membrane-bound organelle that houses the cell’s chromosomes.
Peptides
Peptides are compounds made up of two or more amino acids, which can be the same or different. Peptides are classified according to their length, with dipeptides consisting of two amino acids, tripeptides of three, oligopeptides of two to nine or ten amino acids, polypeptides of ten to ninety-nine or one hundred amino acids, and macropeptides of one hundred amino acids or more being considered proteins.
Polypeptide
Chain of ten or more amino acids held together by peptide bonds.
Polymerase
The protein-making enzyme uses DNA as its template.
PCR (Polymerase Chain Reaction)
In 1985, Kary Mullis devised a method of enzymatically amplifying tiny amounts of DNA to provide enough material for genetic analysis of nucleic acid sequences.
Protein biosynthesis
Translation and transcription are two steps in the protein production process, which takes place on ribosomes inside a cell. Enzymes, hormones, and antibodies are all examples of proteins. Protein is a class of molecules that is predominantly made up of 20 distinct amino acids.
Proteome
Complete set of proteins in a cell, organ, or tissue fluid.
Purine bases
Adenine and guanine are two examples of purine bases.
Pyrimidine bases
DNA and RNA both use the pyrimidine nucleotide uracil, however, RNA uses cytosine and DNA uses mostly thymine.
Restriction enzymes
DNA scissor enzymes are enzymes that detect a particular sequence of letters on DNA and cut the DNA at that sequence.
Ribonucleic acid (RNA)
Ribonucleic acid (RNA) is the “little sister” of deoxyribonucleic acid (DNA), a single-stranded nucleic acid molecule involved in protein production in which the nucleotide uracil (U) replaces thymine.
Ribosome
The ribosome is the cell’s “protein factory,” where proteins are made by reading a copy of a gene.
Telomeres
Normal cells may undergo around 2,000 cell divisions before showing signs of wear and tear, during which time DNA ends (telomeres) that do not carry genetic information shrink.
Transcription
The overwriting of a gene’s DNA into messenger RNA (mRNA).
Translation
The method carried out by ribosomes whereby a protein is synthesized from its constituent amino acids.
Virus
Pathogenic biological structure made up of proteins and nucleic acids that may infect, replicate, and kill host cells.
Viruses are dangerous because they rely on a “host” organism for their metabolism.
Cell
DNA is packed into chromosomes in the cell nucleus, making the cell the smallest reproducing unit in higher animals.